Part F: A Closer Look at....
The Dilution Series and Statistics
Introduction
When you make a dilution series, the tube you start with contains approximately 10^6
cells/mL. From the earlier procedure you know that the suspension actually holds between 1 and
2 million cells/mL, but for now, suppose you have exactly 1 million cells/mL. Since the tube
contains 2 mL, you should have 2 million cells. If you pipet out exactly 0.1 mL, how many cells
will you get? You would expect to get 1/20 of the 2 million cells in the tube, 10^5 cells, but you
probably won't because some extra cells could have been in the 0.1 mL you pulled out. This
expected variation is called the standard deviation, and for our experiment is approximately the
square root of the number of cells you expect to get. Thus, if you expect 105 cells you may only
get within 300 cells of 10^5 (300=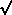 (10^5)).
(10^5)).
Missing a hundred thousand by a mere 300 is not so bad, but notice what happens with
the further dilutions. By the time you expect to get 100 cells, the standard deviation is 10, the
square root of 100. So if you expect to get 100 cells on a plate and in fact you get 90, that is not
necessarily due to poor technique. About 1/3 of the time you should expect to get a result
outside the standard deviation; even getting 80, therefore, may not be the result of poor
technique. Furthermore, just because you get 100 when you expect 100 does not mean you have
great technique; you were also lucky. To develop and demonstrate good technique you must
prepare many dilution series. Try to plate 100 cells 20 times. If your technique is good, you will
get an average of 100 cells per plate, with 13-15 in the 90-110 range, and the rest outside it.
Rules:
- Expected number of cells = (Number in tube) x (fraction of volume withdrawn);
- Standard deviation = square root of expected number of cells;
- Number of plates outside 1 standard deviation ÷ 1/3.
Let's think about designing experiments with these rules in mind. How many cells should
we aim to get on a plate? If we expect to get 100 then our result should be around 100ñ10, while
if we expect to get 10 then we should see around 10ñ3. The uncertainty looks smaller in the
second case. But this is an illusion. 3 is 30% of 10 while 10 is only 10% of 100, so aiming for
100 is smarter than aiming for 10; the relative uncertainty is smaller. What about aiming for
1000 cells on a plate? Then we would expect around 1000ñ+/-30, a 3% relative error. This is true,
the statistical error is smaller, but it is very hard to count 1000 colonies accurately. Using so
many cells introduces a large systematic error into the procedure. When designing an experiment
you have to steer a course between the statistical errors that come with too few cells/plate and the
systematic errors that come with too many cells, or even too many plates; people get tired and
count inaccurately if there is too much to do.
Distributions
Each time you draw 0.1mL from the tube you get some number of cells that you spread
on the plate and that grow up into colonies. We can imagine doing this very many times,
creating a huge number of plates, all made with the same procedure. There are many plates with
100 cells, lots with 107, a few with 74, very few with 13, and so on. This imaginary population
has a distribution of numbers of cells per plate, represented by stacks of plates in the figure
below. Figure 1:
Making a real plate is like pulling a plate out of this ghostly distribution: The plate will probably
come from some where near the mean because there are many more plates there, but it may come
from the wings, that is just less likely.
Real experiments
When we actually do an experiment we are not sure exactly how many cells/mL we have
in suspension, we are not sure that we withdraw exactly 0.1 mL of the suspension, and we do not
how many of the cells are viable (yeast cells eventually die of old age). Thus, although we do
know that we are spreading roughly 100-200 cells on a plate we do not know exactly how many
colonies we should expect. That is, we do not know what distribution our procedure is
producing. We make control plates to figure this out. In the survival experiment, for example,
we keep some plates unexposed to UV radiation. If they grow up to contain 120, 136, and 98
colonies then we can reasonably guess that our procedures have produced a distribution whose
mean is (120+136+98)/3 = 118. The other plates that are exposed to the UV are made by the
same procedures so we expect that before getting any radiation they also come from this
distribution.
Some language from statistics
The example of the dilution series illustrates a number of ideas from statistics, a widely
applicable piece of mathematics. When we use a definite set of procedures to make the plates we
create in our mind's eye a population of plates. The actual control plates are a sample drawn
from the population. The job of statistics is to use the sample to learn things about the
population. The population is characterized by parameters. In this example the mean number of
cells/plate is the parameter we care about. We took our control plates, the sample, and used the
mean of this sample to estimate the mean of the population. We say that the sample mean is an
estimator of the population mean. Our estimation is better with larger samples. If we had made
10 control plates containing 101, 127, 113, 120, 126, 95, 130, 120, 136, 112 cells instead of the
three above our sample size would be 10 instead of 3. We would still estimate the mean to be
118 as we did before but we would feel much more confident of our estimate. We will not go
into the quantitative details of sample size and confidence levels.
The most important characteristics of a sample are its size, its mean and its standard
deviation, or its variance. The size of the sample is usually obvious, and the sample mean is
familiar: it is just the average. The standard deviation and variance of a sample are most easily
explained by an example. Consider the sample of 3 plates above. The mean is 118 so the
deviation of the first plate is 120-118, the deviation of the second is 136-118, and of the third is
98-118. The variance is
((120-118)^2+(136-118)^2+(98-118)^2)/3=242;
that is, the variance is the mean of the squares of the deviations. The standard deviation is just
the square root of the variance, which gives it its other name: the root mean square deviation, or
rms deviation. The standard deviation of a sample is an estimator of the standard deviation of the
population, so in this example we estimate the standard deviation of the population to be 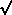 (242)
(242) 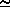 16. This is not far from what we expect from the rule given above (Standard deviation = square
root of expected number of cells):
16. This is not far from what we expect from the rule given above (Standard deviation = square
root of expected number of cells): 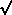 (118) ÷ 11. Why the difference? The rule gives the ideal
standard deviation, to be expected if all procedures are perfectly carried out, which is not
possible in real life. You should be able to get close to that ideal with lots of practice but real
experiments will produce populations with bigger standard deviations than this ideal. Of course,
real samples will sometimes, by chance, have a smaller standard deviation than this ideal! The
sample deviation is just an estimator of the population deviation, it will sometimes be too big and
sometimes too small.
(118) ÷ 11. Why the difference? The rule gives the ideal
standard deviation, to be expected if all procedures are perfectly carried out, which is not
possible in real life. You should be able to get close to that ideal with lots of practice but real
experiments will produce populations with bigger standard deviations than this ideal. Of course,
real samples will sometimes, by chance, have a smaller standard deviation than this ideal! The
sample deviation is just an estimator of the population deviation, it will sometimes be too big and
sometimes too small.
More about distributions
We will discuss several examples of distributions in this section, but the normal
distribution is the most common. (``Normal'' means a particular type of distribution, not an
``ordinary'' distribution.) This is the familiar bell-shaped distribution that describes many
situations, from the number of cells on a plate to students' scores on an exam. It has two
parameters, the mean =  and the standard deviation =
and the standard deviation =  . The mean is the position of the
maximum of the bell-shaped curve and the standard deviation determines the width of the peak.
Consider the qualitative picture sketched below. Both of these distributions have the same mean,
but in the distribution labeled (a) you are unlikely to get a number very far from the mean, while
in the distribution labeled (b) you would not be surprised to get a number quite far from the
mean.
. The mean is the position of the
maximum of the bell-shaped curve and the standard deviation determines the width of the peak.
Consider the qualitative picture sketched below. Both of these distributions have the same mean,
but in the distribution labeled (a) you are unlikely to get a number very far from the mean, while
in the distribution labeled (b) you would not be surprised to get a number quite far from the
mean. Figure 2
Figure 3
Recall that the normal distribution that describes the ideal dilution series experiment done with
perfect technique has standard deviation equal to the square root of the mean, but usually there
are other sources of variation, such as variation in the amount of solution withdrawn for each
plate, so the standard deviation is usually a separate parameter unrelated to the mean. The
mathematical expression for the normal distribution is given below. We give it just for fun; you
don't need to use it now. It uses some symbols you may not yet know, but your teacher, or your
math teacher, can help. The number of plates having x colonies we call N(x) and
N(x)=(1/
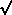 (2*pi
)) exp(-(x-
(2*pi
)) exp(-(x-  )^2/2
)^2/2 ^2)
^2)
One interesting feature of this expression should be noticed: it involves ã, the ratio of the
circumference of a circle to its diameter. Why in the world should geometry have anything to do
with growth of yeast cells? But it does. This is one of the amazing things about mathematics!
The next important distribution in biology is called the Poisson distribution. It is named
for a mathematician who used it. This distribution is involved when we consider mutations
induced by radiation, or the probability of ``cross-over'' when DNA recombines. It also describes
the probability of hearing a ``click'' when you hold a Geiger counter near a radioactive source
and describes the probability of two callers ringing the same phone number at the same time.
What all these situations have in common is that the probability of the interesting event is very
low, but there are many opportunities for the event to occur: it is very unlikely that any particular
cell will mutate, but there are very many cells; it is very unlikely that any particular radioactive
nucleus will decay, but there are very many such nuclei; and so on. We will use the example of
mutation by radiation to explain this distribution.
When you expose the yeast cells that normally make white colonies to UV radiation you
occasionally find a red colony growing up. This can be the result of a mutation. We would like
to know the probability that a single cell will mutate when exposed to this much radiation. Let's
call this p. How can we measure this? The most straightforward idea is probably to expose a lot
of plates, colonies the cells/plate, count the number of mutated cells, and then the number of
mutations divided by the total number of cells exposed is p. So suppose your whole class makes
one thousand plates, with 200 cells/plate, exposes them all to the same amount of radiation, lets
the colonies grow up, and then starts looking for mutants. The result of such an experiment is
sketched below. Figure 4:
Figure 5:
The figure labeled (a) plots the number of plates having n mutations versus n; the figure labeled
(b) plots the probability of having n mutations on a plate versus n. The connection is simple:
there were 903 of the 1000 plates that had 0 mutants so the probability of getting 0 is 0.90, there
were 89 that had 1 mutant so the probability of 1 is 0.09, and so on. We wanted to measure p,
the probability that a single cell will mutate when exposed to this much radiation. The total
number of mutants is 111 (89 plates with one means 89 mutants, 6 plates with two adds twelve,
and so on) and the total number of cells is 1000 plates times 200 cells/plate. Thus we get
p=111/200,000. So we have found out what we wanted to, but there is a little more we can learn.
The dashed line in (b) is a Poisson distribution of the probability of getting n mutants, P(n),
versus n. We can see that this is a pretty good description of the experimental results. The
mathematical expression of the Poisson distribution is
P(n)=(exp - 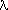 )
) 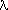 ^)n/n!
^)n/n!
with
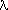 = (p) x (number of cells per plate)
= (p) x (number of cells per plate)
where p is the parameter we wanted. This gives us a different way to determine p. All we need
is P(0), the probability that there are zero mutations on a plate. Because this number is
determined using lots of plates the relative error is small, so it is probably fairly accurate. Here is
how to use P(0) to find "lambda" and then p. From the formula for the Poisson distribution we see that
P(0) equals exp-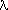 , so
, so 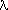 = -ln P(0)= 0.10 and so p=
= -ln P(0)= 0.10 and so p=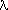 /(number of cells per plate) = 0.10/200
=0.0005, just as we found before.
/(number of cells per plate) = 0.10/200
=0.0005, just as we found before.
The last distribution we will discuss here is the binomial distribution. This gives the
probability of having n successes in N tries. A typical situation is rolling dice or flipping a coin:
the binomial distribution tells the probability of rolling 7 four times in 11 attempts, for example.
A biological example is the mutation experiment we just discussed. We could call ``causing a
mutation'' a success and the number of cells on a plate would be the number of tries: the radiation
tries to mutate each cell and succeeds only with the few mutants.
The binomial distribution is characterized by one parameter, p, the probability of success
on any single try. The mathematical expression of the binomial distribution is this. Let P(n,N)
be the probability of having n successes in N tries. Then if p is the probability of success on any
single try we have
P(n,N)=[N!/n!(N-n)!] x p^n x (1-p)^(N-n).
The quantity inside the square brackets is called "the binomial coefficient" or "N choose n." The
latter name is used because it gives the number of ways to choose n objects from a set of N
things. For example, if you roll two ordinary dice the probability of getting a 7 is 1/6 (can you
figure out why this is so?) so the probability of rolling 7 four times in eleven attempts is (11
choose 4)(1/6)^4(5/6)^7÷ 0.07.
There is a connection between the three distributions we have discussed. If N is very
large then the binomial distribution is practically indistinguishable from the normal distribution
with  = Np and
= Np and  = Np(1-p); if N is very large and p is very small then the binomial distribution
is practically indistinguishable from the Poisson distribution with
= Np(1-p); if N is very large and p is very small then the binomial distribution
is practically indistinguishable from the Poisson distribution with 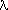 =Np. This equivalence of
Poisson and binomial distributions is the reason we could use the mutation experiment as an
illustration of each of these distributions.
=Np. This equivalence of
Poisson and binomial distributions is the reason we could use the mutation experiment as an
illustration of each of these distributions.
Click here to return
Last updated Wednesday, 04-Dec-2002 14:16:56 CST
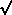 (10^5)).
(10^5)). 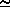 16. This is not far from what we expect from the rule given above (Standard deviation = square
root of expected number of cells):
16. This is not far from what we expect from the rule given above (Standard deviation = square
root of expected number of cells):  and the standard deviation =
and the standard deviation =  . The mean is the position of the
maximum of the bell-shaped curve and the standard deviation determines the width of the peak.
Consider the qualitative picture sketched below. Both of these distributions have the same mean,
but in the distribution labeled (a) you are unlikely to get a number very far from the mean, while
in the distribution labeled (b) you would not be surprised to get a number quite far from the
mean.
. The mean is the position of the
maximum of the bell-shaped curve and the standard deviation determines the width of the peak.
Consider the qualitative picture sketched below. Both of these distributions have the same mean,
but in the distribution labeled (a) you are unlikely to get a number very far from the mean, while
in the distribution labeled (b) you would not be surprised to get a number quite far from the
mean.  )
)