Frequently Asked Questions about GENE and Research Link Yeast Strains
1. Why is the yeast strain (HA1, HA2, HA12, HB1, HB2, HB12) I got
from the supplier not red even though it is supposed to be a red strain?
Answer: The strains are shipped on medium that contains an
excess of adenine. This prevents them from turning red because of
feedback inhibition on the adenine biosynthetic pathway (see answer to
#3, below). It is only when the adenine in the medium is used up
and the yeast attempt to synthesize their own, that they begin to accumulate
the red pigment due to the block in adenine biosyntheis. Streak out
the yeast on YED. It should start to look pink after a couple of
days' growth on YED. If it does not, then you should contact the
supplier.
2. I streaked out the strain from the supplier and I get red
colonies, white colonies, and mixed colonies. What is going on and
which should I pick?
Answer: If this is supposed to be a red strain (e.g.
HA1, HA2, HA12, HB1, HB2, or HB12), then you need to pick a red colony.
What is going on is that white mutants have arisen in the culture and it
is now a mixed culture. By streaking for isolation, you can recover
the pure red strain.
3. Why do white mutants arise in red cultures at such high frequency?
Answer: There is a tremendous disadvantage to the yeast to
make all that red pigment. Red strains will always grow more poorly
than otherwise isogenic white strains. This results in a strong selection
for any mutants that may arise that prevent the red pigment from being
produced. Many of these mutants are secondary mutations that block
an earlier step in adenine biosynthesis than that of the ade1 and
ade2
mutations. This was first shown by Herschel Roman, one of the founding
fathers of yeast genetics, in the 1950's. The pathway looks like
this:
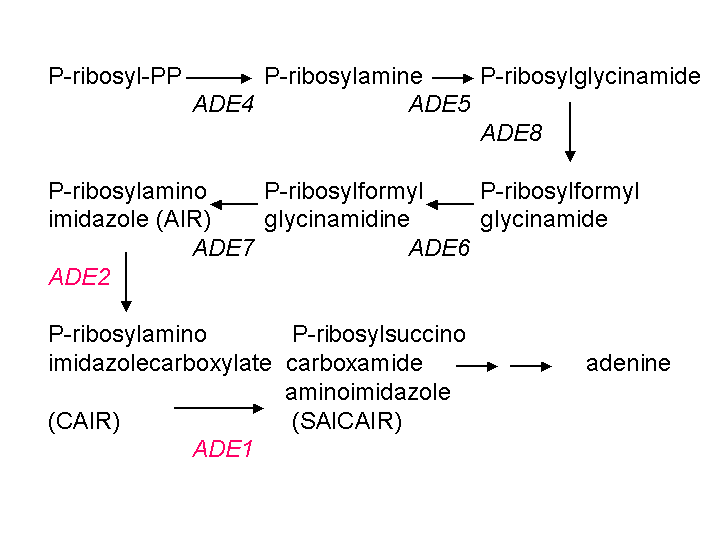 Mutations in ADE1 or ADE2 cause accumulation of the intermediate
AIR, which is converted to form the red pigment. Mutations in any
of the genes earlier in the pathway, such as ADE4, ADE5, ADE8, ADE6,
or
ADE7, block the pathway before the formation of AIR and prevent
accumulation of the pigment. So generally the white mutants contain
two
mutations in the adenine pathway: ade1 or ade2, and
a mutation in one of the genes encoding the enzymes that catalyze an earlier
step. Since there are multiple genes that can be mutated to create
this phenotype, the mutation rate is high.
Mutations in ADE1 or ADE2 cause accumulation of the intermediate
AIR, which is converted to form the red pigment. Mutations in any
of the genes earlier in the pathway, such as ADE4, ADE5, ADE8, ADE6,
or
ADE7, block the pathway before the formation of AIR and prevent
accumulation of the pigment. So generally the white mutants contain
two
mutations in the adenine pathway: ade1 or ade2, and
a mutation in one of the genes encoding the enzymes that catalyze an earlier
step. Since there are multiple genes that can be mutated to create
this phenotype, the mutation rate is high.
4. The yeast strain was red when I got it six months ago but when
I recently tried to grow it up, all I got were white colonies. What
has happened and how do I get the red strain back?
Answer: What has probably happened is that because you kept
the plate around for a long time, you produced conditions that favored
the accumulation of mutations (see question 3, above). Now your culture
mainly consists of double mutants. It may be possible to get the red strain
back by streaking out more colonies--there may be a few single red cells
left--but more than likely you will have to get the strain from the supplier
again.
5. How can I maintain my strains from one school year to another?
Answer:
Short-term storage: Working cultures can be kept on YEPAD agar slants
for 6 months to more than a year, depending on the strain. Non-adenine
requiring strains can be transferred directly to new slants, but adenine-requiring
strains should be purified by isolating single colonies and picking one
that turns pink on YED. (Do not leave on YED more than 3 days.)
Long-term storage: Strains can be stored desiccated in refrigerator
dried onto paper in a suspension of evaporated milk or in ultra-low temperature
refrigerator (Revco) in 15 percent glycerol.
6. Why do HAR and HBR have the same genotypes as HA2 and HB2?
Are they the same strains? And if so, why do they have two different
names?
Answer: They are the same strains, that is, HAR = HA2 and
HBR = HB2. For historical reasons, the different terminologies were
developed for the same strains:
The HA1, HA2, etc., nomenclature is used in the "Two
Genes/One Trait" experiment and the nomenclature is discussed there
in the Teacher Tips for that activity
(Page 31 of Basic Genetics Experiments in the
Classroom Guide to Yeast
Experiments).
The HAO,HAR,HAT notation is used in the "A
Dihybrid Cross Experiment"and the use of the traditional notation is
discussed under Teacher Tips (Page
37 of the Classroom Guide).
Although it now appears later in the book, the Dihybrid
Cross case was developed earlier in the program and the notation was selected
to resemble what teachers were familiar with from Mendelian genetics as
presented in many text books. When Two Genes was developed it was
obviously necessary to distinguish between the two different red strains.
Copyright © 2001, Beth A. Montelone, Ph. D., GENE Project and
Research Link 2000. Last updated 7/10/01.

